Artificial intelligence (AI) is rapidly transforming the landscape of therapeutic antibody discovery and engineering. As the biopharmaceutical industry continues to demand faster, more precise, and cost-effective development pipelines, AI-driven solutions have emerged as game-changers. These methods, particularly those based on deep learning (DL), offer unprecedented capabilities in modeling antibody–antigen (Ab–Ag) interactions, predicting binding affinities, designing novel antibody sequences, and optimizing structural features.
This blog delves into the latest AI-driven approaches in antibody design, exploring how sequence-based, structure-based, and hybrid models are revolutionizing therapeutic development. It also highlights key solutions offered by Creative Biolabs to empower your AI-assisted antibody discovery projects.
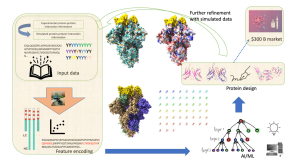
Fig.1 Generalized schematic of DL approaches for Ab sequence design.1
The Challenge of Traditional Antibody Development
Antibody therapeutics, especially monoclonal antibodies (mAbs), have become pivotal in treating cancer, autoimmune diseases, and viral infections. However, their development is a lengthy and costly process. Experimental screening methods like phage and yeast display are limited in throughput—covering up to 10¹⁰ sequences—while the theoretical antibody sequence space exceeds 10¹³ possibilities.
This disparity necessitates smarter strategies for sequence selection, structure modeling, and affinity prediction. Here, AI and DL models step in as powerful tools to compress design cycles and improve accuracy at scale.
The Rise of Deep Learning in Antibody Engineering
DL models are adept at identifying complex nonlinear relationships between biological inputs and outputs. They have already demonstrated success across diverse areas in protein science, including:
-
Predicting 3D protein structures (e.g., AlphaFold, IgFold)
-
Modeling protein–protein interactions
-
Designing antibody CDRs with high affinity
-
Screening sequence libraries for target specificity
Recent advancements have led to the development of sophisticated architectures like recurrent neural networks (RNNs), transformer-based language models, generative adversarial networks (GANs), and diffusion models—all tailored to the unique challenges of antibody biology.
Sequence-Based vs. Structure-Based Approaches: Choosing the Right Strategy
Antibody design methods can be broadly categorized into two types:
Sequence-Based Approaches
These models work with abundant sequence data, predicting antigen-binding potential directly from amino acid sequences. Benefits include:
-
Scalability to billions of sequences
-
Low computational cost
-
Compatibility with metagenomic datasets and synthetic libraries
Models like AntiBERTy, LSTM-based classifiers, and BioPhi fall under this category. They are particularly effective for early-stage screening and in silico humanization.
Structure-Based Approaches
These methods incorporate 3D structural information for greater resolution. They are ideal for:
-
Modeling Ab–Ag docking
-
Predicting structural stability and folding
-
Designing novel paratope–epitope interfaces
DLAB, DeepAb, and IgFold are notable structure-based tools. They are often used in affinity maturation and detailed structural validation stages.
Hybrid Models
Hybrid approaches that combine sequence and structural inputs—such as graph neural networks (GNNs) or autoregressive co-designers—are proving especially effective. These models simulate how local sequence changes influence 3D conformation and binding energetics, allowing for accurate and interpretable design decisions.
Generative Models for Antibody Sequence Design
One of the most exciting areas in DL is generative modeling—where new antibody sequences are created de novo based on learned patterns. This has dramatically advanced the design of CDR loops, especially CDR-H3.
Techniques include:
-
LSTM/GRU models: Predict Ag-specific sequences and distinguish between natural vs. engineered antibodies.
-
VAEs (Variational Autoencoders): Visualize sequence clusters and learn latent features of developability.
-
GANs: Used for large-scale human-like antibody sequence generation.
-
Autoregressive Transformers (e.g., IgLM): Generate full-length antibodies with desired humanization and immunogenicity profiles.
-
Diffusion Models (e.g., AbDiffuser, EvoDiff): Simultaneously optimize structure and sequence through probabilistic modeling.
These models enable rapid design of antibody libraries with higher biophysical relevance and improved functional properties.
Structural Modeling and Screening of Antibody–Antigen Interactions
In addition to sequence generation, AI tools are accelerating the evaluation of binding and developability:
Tools Driving Innovation:
-
DeepAb: Uses crisscross attention and MDS to predict Fv structures and mutation impacts.
-
IgFold: Predicts antibody structures faster than AlphaFold, with high accuracy, even without GPU.
-
ImmuneBuilder (ABodyBuilder2): Offers >100x faster loop prediction with accuracy comparable to traditional tools.
-
DLAB: Employs 3D voxel representations for structure-based virtual screening.
These platforms are particularly valuable in narrowing down candidate sequences before wet-lab validation.
Optimizing Affinity and Developability with AI
Beyond prediction, AI also enables affinity enhancement by proposing targeted mutations in CDRs and framework regions.
For instance, the Binding-ddG-Predictor uses a geometric neural network to rank mutations by their predicted impact on binding energy and structural stability. Surprisingly, many affinity-improving mutations reside outside direct contact regions, making them difficult to identify with traditional methods.
Furthermore, DL-guided developability filters can assess properties such as:
-
Aggregation propensity
-
Thermal and colloidal stability
-
Solubility and post-translational modification sites
These features significantly reduce downstream failure rates in therapeutic development.
Case Study: AI-Guided Discovery Against SARS-CoV-2
The global urgency of the COVID-19 pandemic illustrated the real-world impact of AI in antibody design. Research groups employed DL models to design and validate neutralizing antibodies targeting the SARS-CoV-2 spike protein—achieving in weeks what would have otherwise taken months.
Highlights from these efforts include:
-
Identification of pan-variant CDR mutations via transformer models
-
Prediction of RBD-binding hot spots and escape-resistant residues
-
Development of thermostable antibodies with improved manufacturability
Such outcomes underscore the utility of AI not only in preclinical discovery but also in responding to emerging public health threats with speed and precision.
Empower Your Projects with Creative Biolabs’ AI-Driven Solutions
At Creative Biolabs, we integrate state-of-the-art deep learning into every stage of antibody design. Our proprietary platforms and expert scientists enable precision-guided workflows that deliver results faster and smarter.
Here are three specialized services that can accelerate your next project:
🔹 AI-Augmented Antibody Discovery Services
Leverage generative and predictive models to explore novel antibody sequences and identify high-affinity binders with target specificity. Our platform integrates large-scale sequence databases and unsupervised learning for diversity-driven discovery.
🔹 AI-Based Antibody Screening Services
Rapidly screen millions of antibody candidates using structural and sequence-based prediction engines. Ideal for narrowing down hits before experimental testing.
🔹 AI-Based Antibody Engineering Services
Fine-tune CDRs, optimize humanization, and improve developability parameters through DL-powered simulation and mutation analysis.
Conclusion: A New Era of Intelligent Antibody Design
AI has matured from a computational novelty to a central pillar in antibody engineering. Whether you’re building antibody libraries, improving specificity, or predicting structure–function relationships, deep learning tools offer unmatched speed, precision, and scalability.
As new DL architectures emerge—such as attention-guided diffusion models and MSA-powered language models—the antibody design pipeline will become even more autonomous and efficient.
Ready to revolutionize your antibody development strategy?
Partner with Creative Biolabs and harness the power of AI for next-generation biologics. Contact us today for a custom consultation.
Reference:
1. Kim, Doo Nam, Andrew D. McNaughton, and Neeraj Kumar. “Leveraging artificial intelligence to expedite antibody design and enhance antibody–antigen interactions.” Bioengineering 11.2 (2024): 185. Distributed under the Open Access license CC BY 4.0, without modification.
